purines and pyrimidines are two categories of bases from which nucleosides (base plus ribose sugar) and thus nucleotides (base plus ribose plus phosphate) are made. they are not obtained easily from the diet and as such need to be synthesized de novo. purine synthesis begins with ribosyl-5 phosphate and ATP:
1. ribosyl 5-phosphate and ATP combine to form 5-phosphoribosyl pyrophosphate (PRPP) via PRPP synthetase.
2. the pyrophosphate in PRPP is exchanged for an amine group from glutamine via amido phosphoribosyl transferase, forming 5-phospho ribosylamine.
3. a glycine molecule is added, forming glycinamide ribosyl 5-phosphate.
4. a series of C and N donations: N10 formyl FH4 donates carbon 8.
5. glutamine donates nitrogen 3.
6. N10 formyl FH4 donates carbon 2.
7. aspartate donates carbon 1.
8. CO2 donates carbon 6, forming the purine nucleotide inosine monophosphate (IMP)
IMP can be converted to the nucleotides GMP or AMP as well. the conversion to AMP involves the addition of an aspartate molecule to form adenylosuccinate, using one GTP (in a reaction similar to adding aspartate to citrulline in the urea cycle). adenylosuccinate then loses a fumarate in the subsequent reaction to form AMP. to form GMP, IMP is oxidized by IMP dehydrogenase to xanothine, which is then decarboxylated to GMP with the help of ATP.
regulation of purine synthesis occurs at four places: PRPP synthetase (step 1) is inhibited by ADP and GDP, while amidophosphoribosyl transferase (step 2) is inhibited by AMP and GMP. IMP dehydrogenase and adenylosuccinate synthetase are inhibited by the products they eventually form, GMP and AMP, respectively.
the purine nucleotide cycle occurs in the brain: IMP is converted to AMP by the process described above, and AMP is converted back to IMP by a deaminase reaction, the net result being the conversion of aspartate to fumarate and the production of ammonia (recall from the urea cycle chapter).
 the purine salvage cycle (see diagram) is a system of enzymes that are designed to rebuild the purine nucleotides in the peripheral tissues from the free bases or nucleoside components that are delivered to them. except for adenosine, the general strategy is to convert nucleosides to free bases, and then to nucleotides. there are several types of enzymes involved which interconvert nucleosides, nucleotides, and free bases. 5'nucleotidases convert nucleotides to nucleosides by dephosphorylating the ribose. purine nucleotide phosphorylases cleave the nucleoside into a free base and a phosphorylated ribose. hypoxanthine guanine phosphoribosyl transferase enzymes add an phosphoribosyl unit from PRPP to the base, creating a nucleotide. deaminases can convert AMP to IMP. finally, adenosine kinase can directly phosphorylate adenosine into the nucleotide AMP (it is the only nucleoside that can be converted to a nucleotide directly via salvage enzymes)
the purine salvage cycle (see diagram) is a system of enzymes that are designed to rebuild the purine nucleotides in the peripheral tissues from the free bases or nucleoside components that are delivered to them. except for adenosine, the general strategy is to convert nucleosides to free bases, and then to nucleotides. there are several types of enzymes involved which interconvert nucleosides, nucleotides, and free bases. 5'nucleotidases convert nucleotides to nucleosides by dephosphorylating the ribose. purine nucleotide phosphorylases cleave the nucleoside into a free base and a phosphorylated ribose. hypoxanthine guanine phosphoribosyl transferase enzymes add an phosphoribosyl unit from PRPP to the base, creating a nucleotide. deaminases can convert AMP to IMP. finally, adenosine kinase can directly phosphorylate adenosine into the nucleotide AMP (it is the only nucleoside that can be converted to a nucleotide directly via salvage enzymes)purines can be degraded by salvage enzymes as well: AMP is deaminated to form IMP. the nucleotides IMP and GMP are then converted into nucleosides inosine and guanosine, which are then converted to the bases adenine and guanine. adenine and guanine are converted to xanthosine, which is converted to uric acid and excreted in the urine.
pyrimidine synthesis begins by building the ring structure and adding the phosphoribose later (as opposed to purines, which start with the phosphoribose):
1. glutamine, CO2, and 2ATP combine via carbamoyl phosphate synthetase II to form carbamoyl phosphate.
2. an aspartate is added, forming carbamoyl aspartate, via aspartate transcarbamoylase.
3. dihydroorotase adds an OH, forming dihydroorotate.
4. dihydroorotate is oxidized by dihydroorotate dehydrogenase into orotate, forming the ring structure.
5. a phosphoribosyl unit is added to orotate by orotate phosphoribosyl transferase, forming the nucleotide orotate monophosphate.
6. orotate monophosphate is decarboxylated by OMP decarboxylase to form the nucleotide UMP.
7. UMP can be phosphorylated to UTP.
8. UTP can be converted to the nucleotide CTP.
synthesis of pyrimidines is mainly regulated at the CPS II enzyme level (recall that CPS I is involved in the first step of the urea cycle, in the mitochondria as opposed to the cytosol in pyrimidine synthesis). this enzyme is activated by ATP and inhibited by UTP. pyrimidines are degraded similarly to purines; nucleotides converted to nucleosides converted to free bases cytosine, thiamine, and uracil. cytosine is deaminated to uracil, which is degraded further into CO2, ammonium, and beta-alanine, while thiamine is degraded into CO2, ammonium, and beta-aminoisobutyrate.
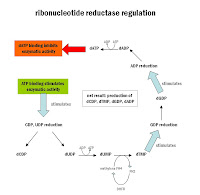 purines and pyrimidines can be converted to the deoxygenated form for use in DNA synthesis. the enzyme that catalyzes this reaction is ribonucleotide reductase, and the electron donor is thioredoxin (which is regenerated by the enzyme thioredoxin reductase and NADPH from the pentose phosphate pathway). ribonucleotide reductase regulation is complex in that it can be activated and inhibited towards specific molecules depending on what binds to it (see diagram).
purines and pyrimidines can be converted to the deoxygenated form for use in DNA synthesis. the enzyme that catalyzes this reaction is ribonucleotide reductase, and the electron donor is thioredoxin (which is regenerated by the enzyme thioredoxin reductase and NADPH from the pentose phosphate pathway). ribonucleotide reductase regulation is complex in that it can be activated and inhibited towards specific molecules depending on what binds to it (see diagram).

No comments:
Post a Comment